You must have Python 3 and Jupyter Notebook already to be installed.
Let`s open your shell and execute next lines:
python3 -m pip install nglview
python3 -m pip install pymatgen
python3 -m pip install ase
jupyter-nbextension enable nglview --py
jupyter notebookThat was enough for me. Now let`s open browser with Jupyter and create new notebook with next code:
# Visualize pymatgen's structure with NGLView https://gist.github.com/lan496/3f60b6474750a6fd2b4237e820fbfea4 https://pymatgen.org/pymatgen.core.structure.html
import nglview
from pymatgen.core import Structure, Lattice
import numpy as np
# https://github.com/pyiron/pyiron/blob/9ba4f1efa57b286d69ee2cdb6b865782eda755f5/pyiron/atomistics/structure/_visualize.py
def plot3d(structure, spacefill=True, show_axes=True):
from itertools import product
from pymatgen.core import Structure
from pymatgen.core.sites import PeriodicSite
eps = 1e-8
sites = []
for site in structure:
species = site.species
frac_coords = np.remainder(site.frac_coords, 1)
for jimage in product([0, 1 - eps], repeat=3):
new_frac_coords = frac_coords + np.array(jimage)
if np.all(new_frac_coords < 1 + eps):
new_site = PeriodicSite(species=species, coords=new_frac_coords, lattice=structure.lattice)
sites.append(new_site)
structure_display = Structure.from_sites(sites)
view = nglview.show_pymatgen(structure_display)
view.add_unitcell()
if spacefill:
view.add_spacefill(radius_type='vdw', radius=0.5, color_scheme='element')
view.remove_ball_and_stick()
else:
view.add_ball_and_stick()
if show_axes:
view.shape.add_arrow([-4, -4, -4], [0, -4, -4], [1, 0, 0], 0.5, "x-axis")
view.shape.add_arrow([-4, -4, -4], [-4, 0, -4], [0, 1, 0], 0.5, "y-axis")
view.shape.add_arrow([-4, -4, -4], [-4, -4, 0], [0, 0, 1], 0.5, "z-axis")
view.camera = "perspective"
return view
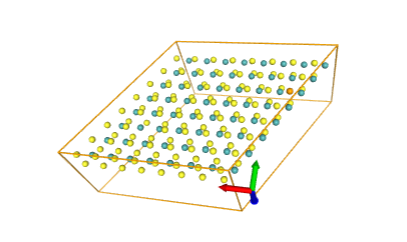
# HERE You need to select your pymatgen json file:
plot3d(
Structure.from_file(
"IDAO-2022-main/data/dichalcogenides_public/structures/6141cf13cc0e69a0cf28ab3b.json"
),
spacefill=True
)After running that You will see interactive nglviewer with structure from your file. В результате должен открыться просмотрщик молекулы nglview. You can scale it, rotate, whatever.
Good luck!